NDFileHDF5
- authors:
Ulrik Pedersen (Diamond Light Source), Arthur Glowacki (Argonne National Laboratory), Alan Greer (Observatory Sciences), Mark Rivers (University of Chicago)
Overview
NDFileHDF5 inherits from NDPluginFile. This plugin uses the HDF5 libraries to store data. HDF5 file format is a self-describing binary format supported by the hdfgroup.
The plugin supports all NDArray datatypes and any number of NDArray dimensions (tested up to 3). It supports storing multiple NDArrays in a single file (in stream or capture modes) where each NDArray gets appended to an extra dimension.
NDArray attributes are stored in the HDF5 file. In case of multi-frame files the attributes are stored in 1D datasets (arrays).
The NDFileHDF5 plugin is created with the NDFileHDF5Configure command, either from C/C++ or from the EPICS IOC shell.
NDFileHDF5Configure (const char *portName, int queueSize, int blockingCallbacks,
const char *NDArrayPort, int NDArrayAddr, size_t maxMemory,
int priority, int stackSize)
For details on the meaning of the parameters to this function refer to the detailed documentation on the NDFileHDF5Configure function in the NDFileHDF5.cpp documentation and in the documentation for the constructor for the NDFileHDF5 class.
File Structure Layout
The HDF5 files comprises a hierachial data structure, similar to a file system structure with directories (groups) and files (datasets) [ref]
XML Defined File Structure Layout
It is possible to define the layout of the data structures in an XML definition file. The XML file allow defining the location of HDF5 Groups, Datasets and Attributes based on detector data (NDArrays), metadata (NDAttributes) and constants (for instance NeXus tags).
The XML definition contains the following 4 main elements: group, dataset, attribute, and hardlink. The terms refer to the HDF5 elements of the same names.
attribute: represent a HDF5 attribute (key, value metadata) and is assigned to groups and datasets
dataset: represent a HDF5 dataset (N-dimensional array). Can contain attributes and is assigned to a group
group: represent a HDF5 group. Groups can contain datasets, attributes and other groups (recursively)
hardlink: represent an HDF5 hard link. Hardlinks can point to other elements in the file, such as datasets. They are analogous to hard links in the Linux file system.
global: Rather than representing a HDF5 object; this element represents a global functionality definition
XML attribute |
Required |
Description |
Value |
|---|---|---|---|
XML “attribute” element attributes |
|||
name |
yes |
Name (key) of the HDF attribute (key, value) pair |
string |
source |
yes |
Definition of where the attribute gets its value from |
Enum string: “constant”, “ndattribute” |
when |
optional |
Event when the attribute data is written |
Enum string: “OnFileOpen” (default), “OnFileClose” |
value |
Required only if source=”constant” |
The constant value to give the attribute |
string (possibly containing an int or float) |
type |
optional - use if source=”constant” |
The constant datatype |
Enum string: “int”, “float”, “string” |
ndattribute |
Required only if source=”ndattribute” |
Name of the areaDetector NDAttribute which is the source of this HDF5 attribute’s data value |
string containing the name of an NDAttribute |
XML “group” element attributes |
|||
name |
yes |
The (relative) name of the HDF5 group |
string |
ndattr_default |
optional |
This attribute flags a group as being a ‘default’ container for NDAttributes which have not been defined to be stored elsewhere. If there is no group defined with ndaddr_default=true, and if the root group does not have auto_ndattr_default=false, then the ‘default’ container will be the root group. |
boolean (default=false) |
auto_ndattr_default |
optional |
If this attribute is present for the root group and is set to false then NDAttributes which have not been defined to be stored elsewhere will not be stored at all |
boolean (default=false) |
XML “dataset” element attributes |
|||
name |
yes |
Name of the HDF5 dataset |
string |
source |
yes |
Definition of where the dataset gets its data values from |
string enum: “detector”, “ndattribute”, “constant” |
when |
optional |
Event when the dataset data is written |
Enum string: “OnFileOpen”, “OnFileClose”, “OnFileWrite” (default) |
value |
Required only if source=”constant” |
Constant value to write directly into the HDF5 dataset |
String - possibly containing int or float values. Arrays of int and float values can also be represented in a comma-separated string |
ndattribute |
Required only if source=”ndattribute” |
The name of the areaDetector NDAttribute to use as a data source for this HDF5 dataset |
string containing the name of the NDAttribute |
det_default |
optional |
Flag to indicate that this HDF5 dataset is the default dataset for the detector to write NDArrays into. Only sensible to set true if source=”detector” |
boolean (default=false) |
XML “global” element attributes |
|||
name |
yes |
Name of the global functionality or parameter to set |
enum string: “detector_data_destination” , (currently only one supported parameter) |
ndattribute |
Required when name=”detector_data_destination” |
Name of the NDAttribute which defines the name of the dataset where incoming NDArrays are to be stored |
string containing the name of an NDAttribute |
XML “hardlink” element attributes |
|||
name |
yes |
Name of the link |
string: string containing the name of the hardlink being created |
target |
yes |
Name of the existing target object in the HDF5 file being linked to. |
string containing the name of the target object being linked to |
An example XML layout file is provided in
ADExample/iocs/simDetectorIOC/iocBoot/iocSimDetector/hdf5_layout_demo.xml.
An XML schema is provided in
ADCore/iocBoot/hdf5_xml_layout_schema.xsd. The schema defines the
syntax that is allowed in the user’s XML definition. It can also be used
with the ‘xmllint’ command to validate a user’s XML definition:
xmllint --noout --schema ADCore/iocBoot/hdf5_xml_layout_schema.xsd /path/to/users/layout.xml
Default File Structure Layout
If no XML Layout Definition file is loaded, the plugin will revert to using its default file structure layout. The default layout is compatible with the plugin’s original, hard-code layout. This layout is actually defined by an XML layout string defined in the source code file NDFileHDF5LayoutXML.cpp.
This default layout is compatible with the Nexus file format. This is achieved by defining a specific hierachial structure of groups and datasets and by tagging elements in the hierachy with certain “NX_class=nnn” attributes. Although Nexus libraries are not used to write the data to disk, this file structure allow Nexus-aware readers to open and read the content of these HDF5 files. This has been tested with the Nexus reader in the GDA application. Hardlinks in the HDF5 file can be used to make the same dataset appear in more than one location. This can be useful for defining a layout that is Nexus compatible, as well as conforming to some other desired layout.
NDArray attributes
The attributes from NDArrays are stored in the HDF5 files. The list of attributes is loaded when a file is opened so XML attributes files should not be reloaded while writing a file in stream mode.
If the dataset is defined in the XML layout file then the user-specified name is used for the dataset. If the dataset is not defined in the XML layout file then the dataset name will be the the NDArray attribute name. The NDArray attribute datasets automatically have 4 HDF “attributes”) to indicate their source type and origin. These are:
NDAttrName: The name of the NDArray attribute.
NDAttrDescrption: The description of the NDArray attribute.
NDAttrSourceType: The source type of the NDArray attribute: NDAttrSourceDriver, NDAttrSourceParam, NDAttrSourceEPICSPV, or NDAttrSourceFunct.
NDAttrSource: The source of the NDArray attribute data, i.e. the name of the EPICS PV, the drvInfo string for the parameter, or the name of the attribute function.
NDArray attributes will be stored as 1D datasets. If the location of an NDArray attribute dataset is not defined in the XML layout file then the dataset will appear in the group that has the property ndattr_default=”true”. If there is no group with that property then the dataset will appear in the root group. If the root group has the property auto_ndattr_default=”false” then datasets that are not explicitly defined in the XML layout file will not appear in the HDF5 file at all.
There are 4 “virtual” attributes that are automatically created. These are:
NDArrayUniqueId: The NDArray.uniqueId value.
NDArrayTimeStamp: The NDArray.timeStamp value.
NDArrayEpicsTSSec: The NDArray.epicsTS.secPastEpoch value.
NDArrayEpicsTSnSec: The NDArray.epicsTS.nsec value.
These properties are added to the property list and will be written to the HDF5 file following the same rules as the actual NDArray ndAttributes described above.
It is possible to validate the syntax of an NDArray attributes XML file.
For example, the command (starting from the iocBoot directory) to
validate the syntax of the
iocBoot/iocSimDetector/simDetectorAttributes.xml file is:
xmllint --noout --schema ./attributes.xsd iocSimDetector/simDetectorAttributes.xml
Default tree structure
The group/dataset structure of the HDF5 files, generated by this plugin:
entry <-- NX_class=NXentry
|
+--instrument <-- NX_class=NXinstrument
|
+--detector <-- NX_class=NXdetector
| |
| +--data <-- NX_class=SDS, signal=1
| |
| +--NDAttributes
| |
| +--ColorMode
|
+--NDAttributes <-- NX_class=NXCollection, ndattr_default="true"
| |
| +--- <-- Any number of NDAttributes from the NDArrays as individual 1D datasets
|
+--performance <-- Performance of the file writing
|
+--timestamp <-- A 2D dataset of different timing measurements taking during file writing
+--data <-- NX_class=NDdata
| |
| +--data <-- Hardlink to /entry/instrument/detector/data
HDF5 File Viewers
Note that if the Single Writer Multiple Reader (SWMR) feature is disabled then it is not possible to do a live “monitoring” of a file which is being written by another process. The file writers mentioned in this section can only be used to open, browse and read the HDF5 files after the NDFileHDF5 plugin has completed writing and closed the file. It is possible to view files while they are being written if the following are all true:
SWMR support is enabled in the plugin.
The viewer client is built using HDF5 1.10 or later.
The file is being accessed using either a local, GPFS, or Lustre file system by both the client and the IOC.
HDFView is a simple GUI tool for viewing and browsing HDF files. It has some limited support for viewing images, plotting graphs and displaying data tables.
The HDF5 libraries also ships with a number of command-line tools for browsing and dumping data.
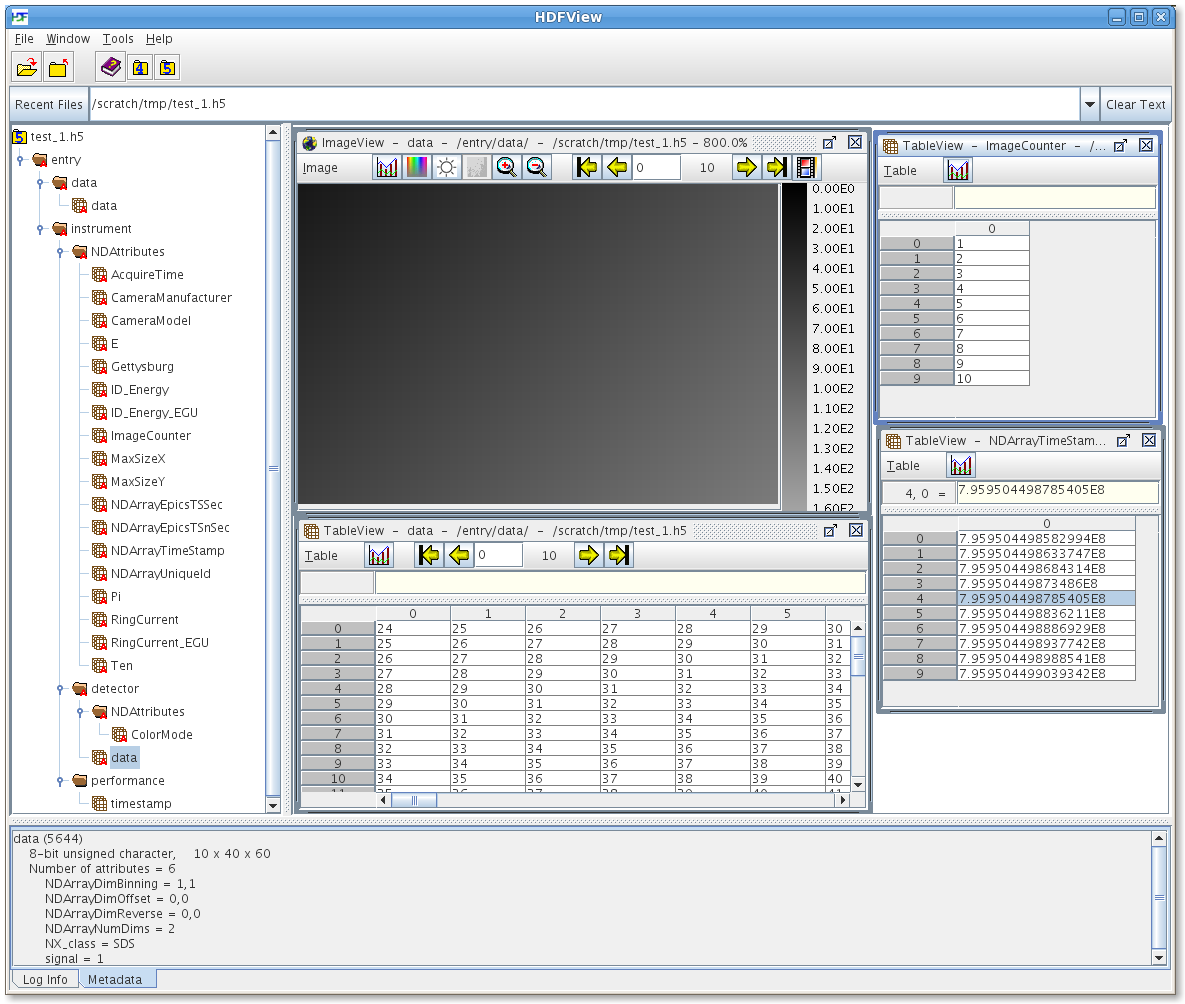
The screenshot below shows the hdfview application with a datafile open. The datafile is generated by the plugin and a number of elements are visible:
The NDArray NDAttributes appear as 1D datasets in the group “/entry/instrument/NDAttributes/”
The image data is in the dataset “/entry/instrument/detector/data”. The metadata (in HDF known as “attributes”) for that dataset indicate 8bit unsigned char data, 10 frames of 60x40 pixels
Image and table view of the first frame data is open
Multiple Dimensions
Both areaDetector and the HDF5 file format supports multidimensional datasets. The dimensions from the NDArray are preserved when writing to the HDF5 file. In multi-frame files (plugin in Stream or Capture mode) an additional dimension is added to the dataset to hold the array of frames.
In addition to the dimensions of the NDArray it is also possible to specify up to 9 extra “virtual” dimensions to store datasets in the file. This is to support applications where a sample is scanned in up to nine dimensions. For two extra dimensions, say X and Y each scan point contains a dataset comprising of multiple frames which can be stored. The length of (i.e. number of points in) each of the virtual dimensions have to be specified before the plugin opens the file for writing. This feature is only supported in the Stream and Capture modes.
This feature allow for creating very large sets of scan data which matches the dimensions of the performed scan in one datafile. Depending on the application this can be a benefit in post processing.
The figure below illustrate the use of the two extra “virtual” dimensions in a 2D (X,Y) raster scan with N frames per point:
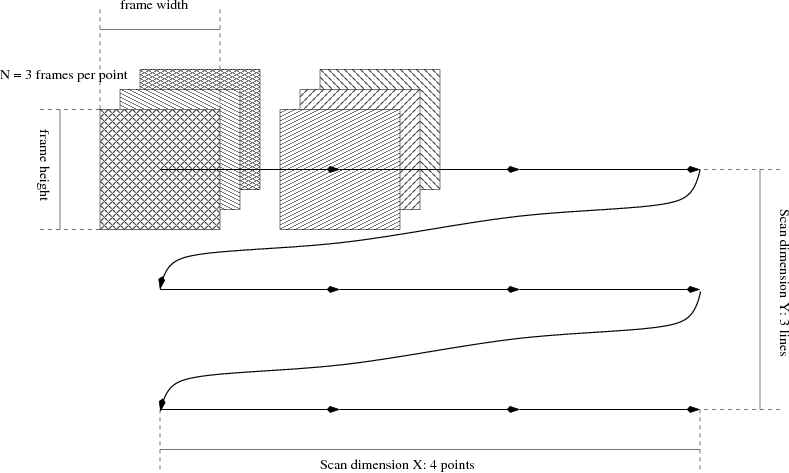
Prior to starting a scan like this the user will need to configure the number of virtual dimensions to use (from 0 up to 9); the number of frames per point; and the length of each of the virtual dimensions (4 x 2 in the example figure). It is not possible to change the number or size of dimensions while the file is open.
For 2D image (greyscale) formats the dimensions in the multiframe HDF5 file are organised as follows. Note that for backwards compatibility (previously only 2 extra dimensions were available) when extra dimensions are specified they are in reverse order 9th,8th,7th,6th,5th,4th,3rd,Yth,Xth and dimensions 2 and 1 are named Y and X:
For a multiframe file with no use of “virtual” dimension the order is: {Nth frame, width, height}
For a multiframe file using 1 “virtual” dimension (X) the order is: {X, Nth frame, width, height}
For a multiframe file using 2 “virtual” dimensions (X,Y) the order is: {Y, X, Nth frame, width, height}
For a multiframe file using 3 “virtual” dimensions (X,Y,3rd) the order is: {3rd, Y, X, Nth frame, width, height}
For a multiframe file using 4 “virtual” dimensions (X,Y,3rd,4th) the order is: {4th, 3rd, Y, X, Nth frame, width, height}
For a multiframe file using 5 “virtual” dimensions (X,Y,3rd,4th,5th) the order is: {5th, 4th, 3rd, Y, X, Nth frame, width, height}
Chunking
This plugin uses HDF5 chunking to store the raw image data. The chunk size (the size of each I/O block) can be either automatically configured (ChunkSizeAuto=Yes) or user-configured (ChunkSizeAuto=No). If automatically configured then the chunk size in each dimension is the size of the NDArray in that dimension. NumFramesChunks defaults to 1, but if it is set higher then each chunk can be multiple NDArrays, (which essentially implies memory caching before writing to disk). Configuring chunking correctly for a given application is a complex matter where both the write performance and the read performance for a given post processing application will have to be evaluated. As a basic starting point, setting the ChunkSizeAuto=Yes, should give a decent result. Further explanations and documentation of the HDF5 chunking feature is available in the HDF5 documentation:
HDF5 documentation advanced topics: Chunking in HDF5
hdfgroup presentation: HDF5 Advanced Topics - Chunking in HDF5
Compression
The HDF5 library supports a number of compression algorithms. When using HDF5 libraries to write and read files the with standard HDF5 compressions (N-bit, szip, and libz) it only need to be switched on when writing and HDF5 enabled applications can read the files without any additional configuration. When using Blosc, LZ4, BSLZ4 and JPEG no additional configuration is required for NDFileHDF5 to write the files, because it registers these compression filters. However, when reading files written with Blosc, LZ4, BSLZ4, or JPEG the environment variable HDF5_PLUGIN_PATH must point to a directory containing the shareable libraries for the decompression filter plugins. This allows any application built with HDF5 1.8.11 or later to read files written with these compression filters. The areaDetector/ADSupport modules builds these shareable libraries for Linux, Windows, and Mac. Only one compression filter can be applied at the time.
The following compression filters are supported in the NDFileHDF5 plugin:
- Lossless SZIP compression is using a separate library from the hdfgroup. NOTE: The szip library contains the following in its COPYING license agreement file:Revocable (in the event of breach by the user or if required by law), royalty-free, nonexclusive sublicense to use SZIP compression software routines and underlying patents for non-commercial, scientific use only is hereby granted by ICs, LLC, to users of and in conjunction with HDF data storage and retrieval file format and software library products.This means that the szip compression should not be used by commercial users without first obtaining a license.
External libz -also lossless
N-bit compression is a bit-packing scheme to be used when a detector provide fewer databits than standard 8,16,32 bit words. Data width and offset in the word is user configurable.
- Blosc compression. Blosc is lossless and contains several compressors,
including LZ4 with Bitshuffle.
LZ4 compression. LZ4 is lossless.
Bitshuffle/LZ4 compression. BSLZ4 is lossless.
JPEG compression. JPEG is lossy, with a user-defined quality factor.
Single Writer Multiple Reader (SWMR)
From version 1-10 of the HDF5 library, reader applications shall be able to access the file whilst it is being written. The plugin has been updated to support the additional SWMR feature when writing a file. The plugin will know if SWMR mode is supported depending on the version of the HDF5 library that the plugin has been compiled against, and SWMR mode can be enabled or disabled by setting the appropriate parameter (disabled by default). Once placed into SWMR mode the plugin accepts parameters to control how often frames are flushed, how often NDAttributes are flushed and the current SWMR status and number of flushes that have taken place are reported for an acquisition. The SWMR active status parameter can be used to signify that it is safe for readers to open the file (the file has been placed into SWMR mode). Data can be flushed to disk on demand using the FlushNow command.
Storing Attributes with Dataset Dimensions
If two “virtual” dimensions are selected with N=3, X=4 and Y=5 then the main dataset will have the dimensions 5x4x3x600x800.
If the NDAttribute datasets are stored in the standard way then the dataset would be a single dimension array of 60 items.
If however, the NDAttribute dataset is stored with the same dimensionality as the main dataset then the NDAttribute dataset will have the dimensions of 5x4x3.
Selective Positional Placement of Individual Frames
The plugin can now store frames at specified positions within a dataset. Each individual dimension index can be specified with NDAttributes that are attached to the frame. The names of the NDAttributes that will be used as the index values for each dimension are specified using parameters (see parameter table below). The NDAttribute values themselves must be integer type zero based index values; if values are specified that are outside the maximum size of the dataset in any dimension then the acquisition will fail. A named attribute must be specified for each additional dimension if position placement mode is to be used, if attributes are not specified or not named correctly then the acquisition will fail.
The position placement mode of operation is enabled by setting the PositionMode parameter to “On” and can be used with or without SWMR mode enabled.
Writing Index Datasets
The plugin will write out an index dataset for an extra dimension multidimensional dataset if requested. This writing out of index values can only take place when running in selective positional placement mode and when storing attributes with dataset dimensions (see above). The index parameters take the name of the NDAttribute that contains the index values for the particular dimensions that you are interested in.
For example, extra X and Y dimensions specified along with positional placement mode looking for NDAttributes called “x” and “y”. If the index dataset for the X dimension is set to “x” also then an additional dataset will be written that contains only the index values for the X dimension. The dataset will be a 1 dimensional dataset. Consider the following index values
x=0, y=0
x=1, y=0
x=0, y=1
x=1, y=1
x=0, y=2
x=1, y=2
If the X index parameter is set to x then a 1D dataset will be produced containing the values (0, 1). If the Y index parameter is set to y then a 1D dataset will be produced containing the values (0, 1, 2).
Parameters and Records
Parameter Definitions and EPICS Record Definitions in NDFileHDF5.template |
|||||
|---|---|---|---|---|---|
asyn interface |
Access |
Description |
drvInfo string |
EPICS record name |
EPICS record type |
HDF5 XML Layout Definition |
|||||
asynOctet |
r/w |
XML filename, pointing to an XML HDF5 Layout Definition, This waveform also supports loading raw XML code directly; up to a maximum of 1MB long (NELM=1MB) |
HDF5_layoutFilename |
$(P)$(R)XMLFileName, $(P)$(R)XMLFileName_RBV |
waveform |
asynInt32 |
r/o |
Flag to report the validity (xml syntax only) of the loaded XML. Updated when the XMLFileName is updated with a new filename and when the XML file is read at HDF5 file creation |
HDF5_layoutValid |
$(P)$(R)XMLValid_RBV |
bi |
asynOctet |
r/o |
XML parser error message |
HDF5_layoutErrorMsg |
$(P)$(R)XMLErrorMsg_RBV |
waveform |
HDF5 Chunk Configuration |
|||||
asynInt32 |
r/w |
No (0) or Yes (1). If Yes then the chunk size for each dimension of the NDArray is set to be the size of the NDArray in that dimension. |
HDF5_chunkSizeAuto |
$(P)$(R)ChunkSizeAuto, $(P)$(R)ChunkSizeAuto_RBV |
bo, bi |
asynInt32 |
r/w |
Configure HDF5 “chunking” to approriate size for the filesystem: sets number of columns (dimension 0 of NDArray) to use per chunk |
HDF5_nColChunks |
$(P)$(R)NumColChunks, $(P)$(R)NumColChunks_RBV |
longout, longin |
asynInt32 |
r/w |
Configure HDF5 “chunking” to approriate size for the filesystem: sets number of rows (dimension 1 of NDArray) to use per chunk |
HDF5_nRowChunks |
$(P)$(R)NumRowChunks, $(P)$(R)NumRowChunks_RBV |
longout, longin |
asynInt32 |
r/w |
Configure HDF5 “chunking” to approriate size for the filesystem: sets the number of elements in dimension N use per chunk |
HDF5_chunkSize |
$(P)$(R)ChunkSize(N), $(P)$(R)ChunkSize(N)_RBV |
longout, longin |
asynInt32 |
r/w |
Configure HDF5 “chunking” to approriate size for the filesystem: sets number of NDArrays to use per chunk. Setting this parameter > 1 essentially implies using in-memory cache as HDF5 only writes full chunks to disk. |
HDF5_nFramesChunks |
$(P)$(R)NumFramesChunks, $(P)$(R)NumFramesChunks_RBV |
longout, longin |
Disk Boundary Alignment |
|||||
asynInt32 |
r/w |
Set the disk boundary alignment in bytes. This parameter can be used to optimise file I/O performance on some file systems. For instance on the Lustre file system where the it is optimal to align data to the ‘stripe size’ (default 1MB). , This parameter applies to all datasets in the file. , Setting this parameter to 0 disables use of disk boundary alignment. , Warning: setting this parameter to a larger size than the size of a single chunk will cause datafiles to grow larger than the actual contained data. |
HDF5_chunkBoundaryAlign |
$(P)$(R)BoundaryAlign, $(P)$(R)BoundaryAlign_RBV |
longout, longin |
asynInt32 |
r/w |
Set a minimum size (bytes) of chunk or dataset where boundary alignment is to be applied. This can be used to filter out small datasets like NDAttributes from the boundary alignment as it could blow up the file size. , Setting this parameter to 0 will disable the use of boundary alignment |
HDF5_chunkBoundaryThreshold |
$(P)$(R)BoundaryThreshold, $(P)$(R)BoundaryThreshold_RBV |
longout, longin |
Metadata |
|||||
asynInt32 |
r/w |
Enable or disable support for storing NDArray attributes in file |
HDF5_storeAttributes |
$(P)$(R)StoreAttr, $(P)$(R)StoreAttr_RBV |
bo, bi |
asynInt32 |
r/w |
Enable or disable support for storing file IO timing measurements in file |
HDF5_storePerformance |
$(P)$(R)StorePerform, $(P)$(R)StorePerform_RBV |
bo, bi |
asynInt32 |
r/w |
Turn on or off NDAttribute dataset dimensions (1 = On, 0 = Off). When switched on NDAttribute datasets are forced to have the same dimensionality as the main dataset. |
HDF5_dimAttDatasets |
$(P)$(R)DimAttDatasets, $(P)$(R)DimAttDatasets_RBV |
bo, bi |
asynFloat64 |
r/w |
Fill value for the dataset to be set in its creation property list. |
HDF5_fillValue |
$(P)$(R)FillValue, $(P)$(R)FillValue_RBV |
ao, ai |
SWMR |
|||||
asynInt32 |
r/o |
Does the HDF5 library version support SWMR mode of operation (1 = Supported, 0 = Not Supported). |
HDF5_SWMRSupported |
$(P)$(R)SWMRSupported_RBV |
bi |
asynInt32 |
r/w |
Turn on or off SWMR mode for the next acquisition (1 = On, 0 = Off). Turning on will only work if SWMR mode is supported. |
HDF5_SWMRMode |
$(P)$(R)SWMRMode, $(P)$(R)SWMRMode_RBV |
bo, bi |
asynInt32 |
r/o |
This value is set to 1 once a file has been opened and placed into SWMR mode. It returns to 0 once the acquisition has completed. |
HDF5_SWMRRunning |
$(P)$(R)SWMRActive_RBV |
bi |
asynInt32 |
r/w |
Flush the image to disk every N’th frame. |
HDF5_flushNthFrame |
$(P)$(R)NumFramesFlush, $(P)$(R)NumFramesFlush_RBV |
longout, longin |
asynInt32 |
r/w |
This value is used to determine when to flush NDAttribute datasets to disk, and the corresponding datasets chunk size. A value of zero will default where possible to the size of the dataset for a one dimensional dataset. |
HDF5_NDAttributeChunk |
$(P)$(R)NDAttributeChunk, $(P)$(R)NDAttributeChunk_RBV |
longout, longin |
asynInt32 |
r/o |
The number of flushes that have taken place for the current acquisition. In the case where flushing occurs for every frame and no flushing is set for NDAttribute datasets then this value will increment by one for each frame, and once the acquisition has completed it will jump by the number of flushes required for the NDAttribute datasets, one flush for each dataset. |
HDF5_SWMRCbCounter |
$(P)$(R)SWMRCbCounter_RBV |
longin |
asynInt32 |
r/w |
Forces an immediate HDF5 flush. |
HDF5_SWMRFlushNow |
$(P)$(R)FlushNow |
busy |
Additional Virtual Dimensions |
|||||
asynInt32 |
r/w |
Number of extra dimensions [0..9] |
HDF5_nExtraDims |
$(P)$(R)NumExtraDims, $(P)$(R)NumExtraDims_RBV |
mbbo, mbbi |
asynInt32 |
r/w |
Size of extra dimension N (no. of frames per point) |
HDF5_extraDimSizeN |
$(P)$(R)ExtraDimSizeN, $(P)$(R)ExtraDimSizeN_RBV |
|
asynOctet |
r/o |
HDF5_extraDimNameN |
Name of extra dimension N |
$(P)$(R)ExtraDimNameN_RBV |
stringin |
asynInt32 |
r/w |
Size of extra dimension X |
HDF5_extraDimSizeX |
$(P)$(R)ExtraDimSizeX, $(P)$(R)ExtraDimSizeX_RBV |
longout, longin |
asynOctet |
r/o |
Name of extra dimension X |
HDF5_extraDimNameX |
$(P)$(R)ExtraDimNameX_RBV |
stringin |
asynInt32 |
r/w |
Chunk of extra dimension X |
HDF5_extraDimChunkX |
$(P)$(R)ExtraDimChunkX, $(P)$(R)ExtraDimChunkX_RBV |
longout, longin |
asynInt32 |
r/w |
Size of extra dimension Y |
HDF5_extraDimSizeY |
$(P)$(R)ExtraDimSizeY, $(P)$(R)ExtraDimSizeY_RBV |
longout, longin |
asynOctet |
r/o |
Name of extra dimension Y |
HDF5_extraDimNameY |
$(P)$(R)ExtraDimNameY_RBV |
stringin |
asynInt32 |
r/w |
Chunk of extra dimension Y |
HDF5_extraDimChunkY |
$(P)$(R)ExtraDimChunkY, $(P)$(R)ExtraDimChunkY_RBV |
longout, longin |
asynInt32 |
r/w |
Size of the K’th extra dimension, for K in [3..9] |
HDF5_extraDimSize<K> |
$(P)$(R)ExtraDimSize<K>, $(P)$(R)ExtraDimSize<K>_RBV E.g. $(P)$(R)ExtraDimSize3_RBV |
longout, longin |
asynOctet |
r/o |
Name of the K’th extra dimension, for K in [3..9] |
HDF5_extraDimName<K> |
$(P)$(R)ExtraDimName<K>_RBV E.g $(P)$(R)ExtraDimName8_RBV |
stringin |
asynInt32 |
r/w |
Chunk of the K’th extra dimension, for K in [3..9] |
HDF5_extraDimChunk<K> |
$(P)$(R)ExtraDimChunk<K>, $(P)$(R)ExtraDimChunk<K>_RBV E.g. $(P)$(R)ExtraDimChunk9 |
longout, longin |
Positional Placement |
|||||
asynInt32 |
r/w |
Turn on/off positional placement mode |
HDF5_posRunning |
$(P)$(R)PositionMode, $(P)$(R)PositionMode_RBV |
bo, bi |
asynOctet |
r/w |
Specify the NDAttribute name for the N index |
HDF5_posNameDimN |
$(P)$(R)PosNameDimN, $(P)$(R)PosNameDimN_RBV |
stringout, stringin |
asynOctet |
r/w |
Specify the NDAttribute name for the X index |
HDF5_posNameDimX |
$(P)$(R)PosNameDimX, $(P)$(R)PosNameDimX_RBV |
stringout, stringin |
asynOctet |
r/w |
Specify the NDAttribute name for the Y index |
HDF5_posNameDimY |
$(P)$(R)PosNameDimY, $(P)$(R)PosNameDimY_RBV |
stringout, stringin |
asynOctet |
r/w |
Specify the NDAttribute name for the K’th index, for K in [3..9] |
HDF5_posNameDim<K> |
$(P)$(R)PosNameDim<K>, $(P)$(R)PosNameDim<K>_RBV E.g. $(P)$(R)PosNameDim5 |
stringout, stringin |
Index Datasets |
|||||
asynOctet |
r/w |
Specify the NDAttribute index for the N dimension |
HDF5_posIndexDimN |
$(P)$(R)PosIndexDimN, $(P)$(R)PosIndexDimN_RBV |
stringout, stringin |
asynOctet |
r/w |
Specify the NDAttribute index for the X dimension |
HDF5_posIndexDimX |
$(P)$(R)PosIndexDimX, $(P)$(R)PosIndexDimX_RBV |
stringout, stringin |
asynOctet |
r/w |
Specify the NDAttribute index for the Y dimension |
HDF5_posIndexDimY |
$(P)$(R)PosIndexDimY, $(P)$(R)PosIndexDimY_RBV |
stringout, stringin |
asynOctet |
r/w |
Specify the NDAttribute index for the K’th dimension, for K in [3..9] |
HDF5_posIndexDim<K> |
$(P)$(R)PosIndexDim<K>, $(P)$(R)PosIndexDim<K>_RBV E.g. $(P)$(R)PosIndexDim9_RBV |
stringout, stringin |
Runtime Statistics |
|||||
asynFloat64 |
r/o |
Total runtime in seconds from first frame to file closed |
HDF5_totalRuntime |
$(P)$(R)RunTime |
ai |
asynFloat64 |
r/o |
Overall IO write speed in megabit per second from first frame to file closed |
HDF5_totalIoSpeed |
$(P)$(R)IOSpeed |
ai |
Compression Filters |
|||||
asynInt32 |
r/w |
Select or switch off compression filter. Choices are: [None, N-bit, szip, zlib, Blosc, BSLZ4, LZ4, JPEG] |
HDF5_compressionType |
$(P)$(R)Compression, $(P)$(R)Compression_RBV |
mbbo, mbbi |
asynInt32 |
r/w |
N-bit compression filter: number of data bits per pixel |
HDF5_nbitsPrecision |
$(P)$(R)NumDataBits, $(P)$(R)NumDataBits_RBV |
longout, longin |
asynInt32 |
r/w |
N-bit compression filter: dataword bit-offset in pixel |
HDF5_nbitsOffset |
$(P)$(R)DataBitsOffset, $(P)$(R)DataBitsOffset_RBV |
longout, longin |
asynInt32 |
r/w |
szip compression filter: number of pixels in filter [1..32] |
HDF5_szipNumPixels |
$(P)$(R)SZipNumPixels, $(P)$(R)SZipNumPixels_RBV |
longout, longin |
asynInt32 |
r/w |
zlib compression filter: compression level [1..9] |
HDF5_zCompressLevel |
$(P)$(R)ZLevel, $(P)$(R)ZLevel_RBV |
longout, longin |
asynInt32 |
r/w |
Blosc compressor. Choices are: [BloscLZ, LZ4, LZ4HC, SNAPPY, ZLIB, ZSTD] |
HDF5_bloscCompressor |
$(P)$(R)BloscCompressor, $(P)$(R)BloscCompressor_RBV |
mbbo, mbbi |
asynInt32 |
r/w |
Blosc shuffle. Choices are: [None, Byte, Bit] |
HDF5_bloscShuffle |
$(P)$(R)BloscShuffle, $(P)$(R)BloscShuffle_RBV |
mbbo, mbbi |
asynInt32 |
r/w |
Blosc compression filter: compression level [0..9] |
HDF5_bloscCompressLevel |
$(P)$(R)BloscLevel, $(P)$(R)BloscLevel_RBV |
longout, longin |
asynInt32 |
r/w |
JPEG quality level [1..100] |
HDF5_jpegQuality |
$(P)$(R)JPEGQuality, $(P)$(R)JPEGQuality_RBV |
longout, longin |
Screenshots
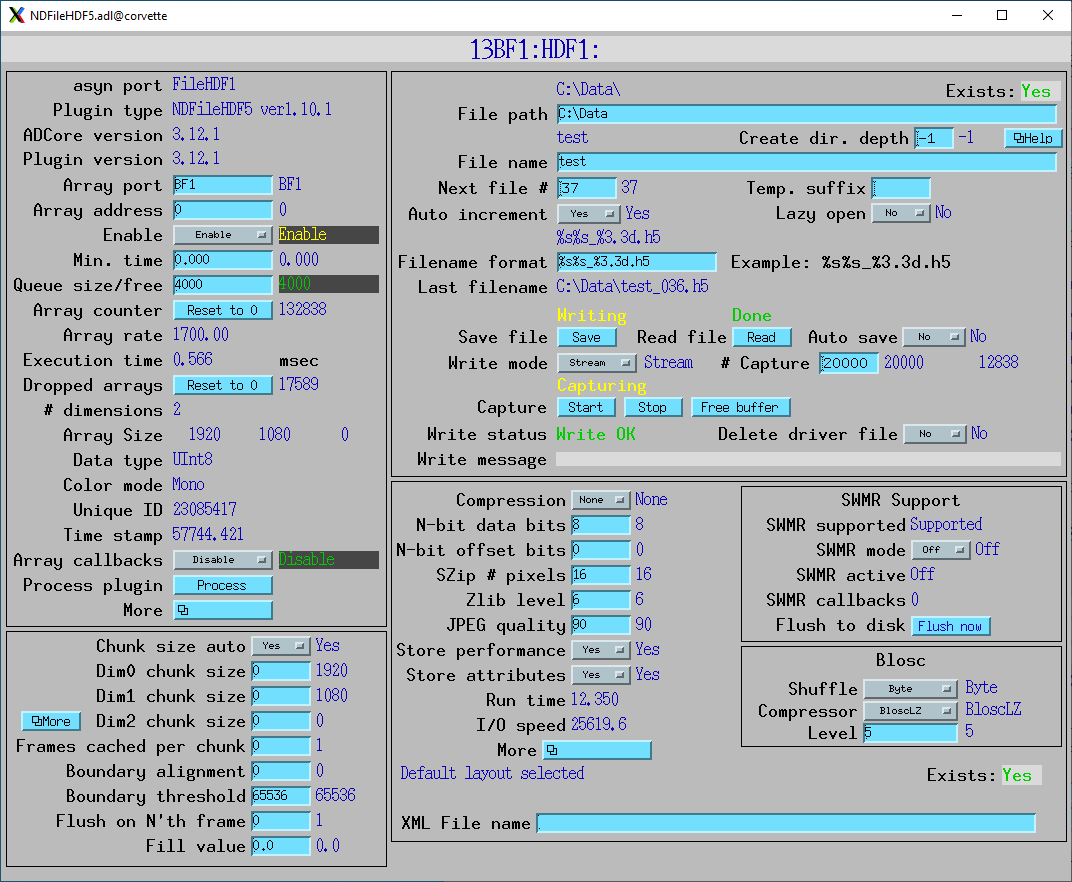
NDFileHDF5.adl